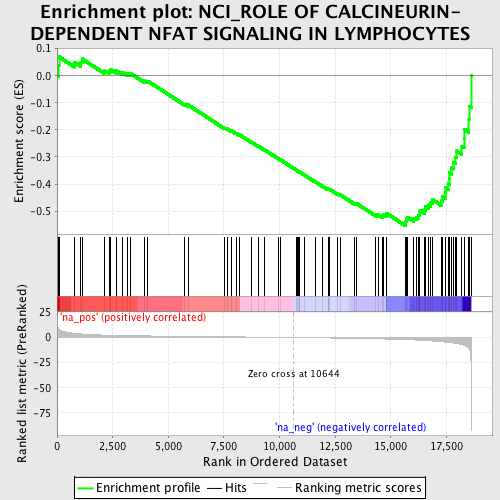
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
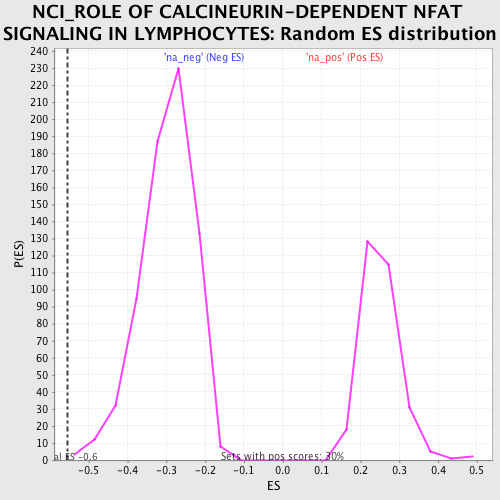
| Upregulated in class | na_neg |
| GeneSet | NCI_ROLE OF CALCINEURIN-DEPENDENT NFAT SIGNALING IN LYMPHOCYTES |
| Enrichment Score (ES) | -0.55462724 |
| Normalized Enrichment Score (NES) | -1.8593457 |
| Nominal p-value | 0.0014285714 |
| FDR q-value | 0.06567158 |
| FWER p-Value | 0.704 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 45 | 9.359 | 0.0381 | No | ||
| 2 | CDK4 | 86 | 7.778 | 0.0697 | No | ||
| 3 | BAD | 784 | 4.120 | 0.0500 | No | ||
| 4 | EGR3 | 1068 | 3.569 | 0.0502 | No | ||
| 5 | IL2RA | 1123 | 3.488 | 0.0624 | No | ||
| 6 | PRKACA | 2114 | 2.419 | 0.0195 | No | ||
| 7 | IL4 | 2334 | 2.280 | 0.0175 | No | ||
| 8 | FKBP8 | 2409 | 2.232 | 0.0232 | No | ||
| 9 | SLC3A2 | 2671 | 2.093 | 0.0182 | No | ||
| 10 | MAPK3 | 2925 | 1.958 | 0.0131 | No | ||
| 11 | RNF128 | 3148 | 1.861 | 0.0091 | No | ||
| 12 | YWHAQ | 3313 | 1.791 | 0.0081 | No | ||
| 13 | CALM1 | 3948 | 1.579 | -0.0193 | No | ||
| 14 | E2F1 | 4079 | 1.545 | -0.0196 | No | ||
| 15 | TBX21 | 5746 | 1.112 | -0.1047 | No | ||
| 16 | PRKCZ | 5893 | 1.076 | -0.1079 | No | ||
| 17 | PTPN1 | 7537 | 0.714 | -0.1934 | No | ||
| 18 | PRKCA | 7649 | 0.691 | -0.1964 | No | ||
| 19 | XPO1 | 7829 | 0.653 | -0.2032 | No | ||
| 20 | RAN | 8046 | 0.606 | -0.2122 | No | ||
| 21 | NR4A1 | 8191 | 0.573 | -0.2175 | No | ||
| 22 | PPARG | 8728 | 0.461 | -0.2444 | No | ||
| 23 | IL2 | 9059 | 0.390 | -0.2605 | No | ||
| 24 | CSF2 | 9327 | 0.329 | -0.2735 | No | ||
| 25 | CASP3 | 9967 | 0.184 | -0.3072 | No | ||
| 26 | PRKCE | 10026 | 0.168 | -0.3096 | No | ||
| 27 | FOS | 10770 | -0.029 | -0.3495 | No | ||
| 28 | IL3 | 10817 | -0.043 | -0.3518 | No | ||
| 29 | TNF | 10851 | -0.051 | -0.3534 | No | ||
| 30 | YWHAZ | 10875 | -0.059 | -0.3544 | No | ||
| 31 | EGR4 | 10880 | -0.060 | -0.3543 | No | ||
| 32 | PRKCH | 11098 | -0.114 | -0.3655 | No | ||
| 33 | FOSL1 | 11626 | -0.268 | -0.3928 | No | ||
| 34 | PTGS2 | 11918 | -0.354 | -0.4070 | No | ||
| 35 | CD40LG | 12191 | -0.434 | -0.4198 | No | ||
| 36 | PRKCD | 12195 | -0.435 | -0.4180 | No | ||
| 37 | NFATC2 | 12258 | -0.453 | -0.4194 | No | ||
| 38 | YWHAE | 12605 | -0.561 | -0.4356 | No | ||
| 39 | JUN | 12736 | -0.603 | -0.4400 | No | ||
| 40 | MEF2D | 13381 | -0.847 | -0.4711 | No | ||
| 41 | IFNG | 13438 | -0.868 | -0.4704 | No | ||
| 42 | CAMK4 | 14327 | -1.247 | -0.5128 | No | ||
| 43 | SFN | 14447 | -1.307 | -0.5136 | No | ||
| 44 | CSNK2A1 | 14622 | -1.386 | -0.5170 | No | ||
| 45 | CTLA4 | 14652 | -1.403 | -0.5125 | No | ||
| 46 | YWHAB | 14792 | -1.481 | -0.5135 | No | ||
| 47 | CSNK1A1 | 14797 | -1.485 | -0.5073 | No | ||
| 48 | YWHAG | 15675 | -2.080 | -0.5456 | Yes | ||
| 49 | CBLB | 15678 | -2.083 | -0.5367 | Yes | ||
| 50 | CREBBP | 15702 | -2.106 | -0.5288 | Yes | ||
| 51 | KPNB1 | 15749 | -2.141 | -0.5220 | Yes | ||
| 52 | IRF4 | 16037 | -2.419 | -0.5270 | Yes | ||
| 53 | PTPRK | 16140 | -2.524 | -0.5216 | Yes | ||
| 54 | CALM2 | 16239 | -2.624 | -0.5155 | Yes | ||
| 55 | GATA3 | 16277 | -2.658 | -0.5059 | Yes | ||
| 56 | KPNA2 | 16310 | -2.686 | -0.4960 | Yes | ||
| 57 | FKBP1A | 16523 | -2.958 | -0.4946 | Yes | ||
| 58 | MAPK8 | 16556 | -2.993 | -0.4834 | Yes | ||
| 59 | MAPK14 | 16698 | -3.190 | -0.4772 | Yes | ||
| 60 | JUNB | 16800 | -3.351 | -0.4681 | Yes | ||
| 61 | POU2F1 | 16867 | -3.457 | -0.4567 | Yes | ||
| 62 | YWHAH | 17289 | -4.152 | -0.4614 | Yes | ||
| 63 | MAP3K8 | 17320 | -4.218 | -0.4447 | Yes | ||
| 64 | PIM1 | 17464 | -4.569 | -0.4326 | Yes | ||
| 65 | CALM3 | 17473 | -4.587 | -0.4132 | Yes | ||
| 66 | GSK3B | 17591 | -4.835 | -0.3985 | Yes | ||
| 67 | FASLG | 17629 | -4.926 | -0.3792 | Yes | ||
| 68 | PRKCQ | 17644 | -4.969 | -0.3584 | Yes | ||
| 69 | ITCH | 17735 | -5.234 | -0.3405 | Yes | ||
| 70 | MAP3K1 | 17806 | -5.436 | -0.3208 | Yes | ||
| 71 | FOXP3 | 17901 | -5.743 | -0.3009 | Yes | ||
| 72 | MAPK9 | 17946 | -5.900 | -0.2777 | Yes | ||
| 73 | EGR1 | 18190 | -7.067 | -0.2602 | Yes | ||
| 74 | NFATC1 | 18298 | -7.855 | -0.2319 | Yes | ||
| 75 | NFATC3 | 18304 | -7.953 | -0.1977 | Yes | ||
| 76 | DGKA | 18512 | -10.953 | -0.1614 | Yes | ||
| 77 | BCL2 | 18517 | -11.183 | -0.1132 | Yes | ||
| 78 | EGR2 | 18610 | -27.324 | 0.0003 | Yes |